Main Page
From PileLine
Contents |
What's new
PileLine GUI 1.3 has been released on July 29, 2011
The major changes in this version are:
- Compatible with the latest pileline tools (version 1.2).
- GP Files viewer: using JXTable instead of JTable to show data rows and added a new functionality to set the number of rows to retrieve by the seeker.
- Genome Browser:
- Added support to 2smc output Pileup files.
- File loader.
- Using double-buffer for each track.
- Allow the user to center the sequence in a given position.
- Several bug fixes in genome browser.
- Bugfix when joinning files with rightouter/leftouter options.
PileLine 1.2 has been released on July 29, 2011
The major changes in this version are:
- pileline-nsmc
- Added two new parameters: -c (column to include when using -e), --ref-col and --allele-col for reference genome and alternative (allele) columns.
- pileline-fulljoin:
- Support for different seq-col and pos-col for each input file via --seq-cols and --pos-cols
- pileline-fastjoin:
- Bugfix. NullPointerException when using custom columns and the sequence or position are the last column.
- Added new behaviour: The data of each joined file now corresponds to all columns but those of sequence and position.
- pileline-fastseek:
- Bugfix. Hang when using a custom --seq-col
- pileline-2smc, pileline-nsmc:
- Added column names in output files as a comment.
- core:
- Bugfix in tabix-based intervals index. Exception "Cannot getOverlappingIntervals with an iteration in progress" is thrown when getting overlapping intervals and getting only the first. This also produces that unit test didn't complete successfully.
PileLine 1.1 has been released on Feb 11, 2011
The major changes in this version are:
- New pileline-fulljoin command to join n GP files.
- New pileline-nsmc mode: intervals mode. Now it can check the reproducibility of mutations by entire intervals (i.e.:genes, provided as .bed), instead of only exact genome position.
- Added BGZF compatibility. This is the samtools compressed format of GP files. All PileLine commands are adapted to receive both bgz-compressed or uncompressed files, transparently. If you want to know how to compress your GP files, please see: Compress/index input with bgzip+tabix
- Added .fai compatibility for genome indexes (needed in pileline-genotest). The pileline-genindex command is now deprecated. You should use the "samtools faidx" command instead.
- Added more flexibility to include custom GP files, by indicating the sequence, start and stop columns in files.
- Bugfix: Bed files is treated in the standard form: (0-based and the last position in the interval is excluded).
- Bugfix: Prevent fastseek from hanging due to bad input files (i.e.: zip compressed, non-ascii, non tab-separated). A small heuristic check is included before processing the file.
Please note: we are updating the commands help wiki pages
Welcome to PileLine Wiki
PileLine is a flexible command-line toolkit for efficient handling, filtering, and comparison of genomic position (GP) files produced by next-generation sequencing experiments (i.e. pileup, BED,GFF, or VCF files). PileLine is designed to be memory efficient by performing on-disk operations over sorted GP files directly.
PileLine is available for downloading at: http://sourceforge.net/projects/pilelinetools/
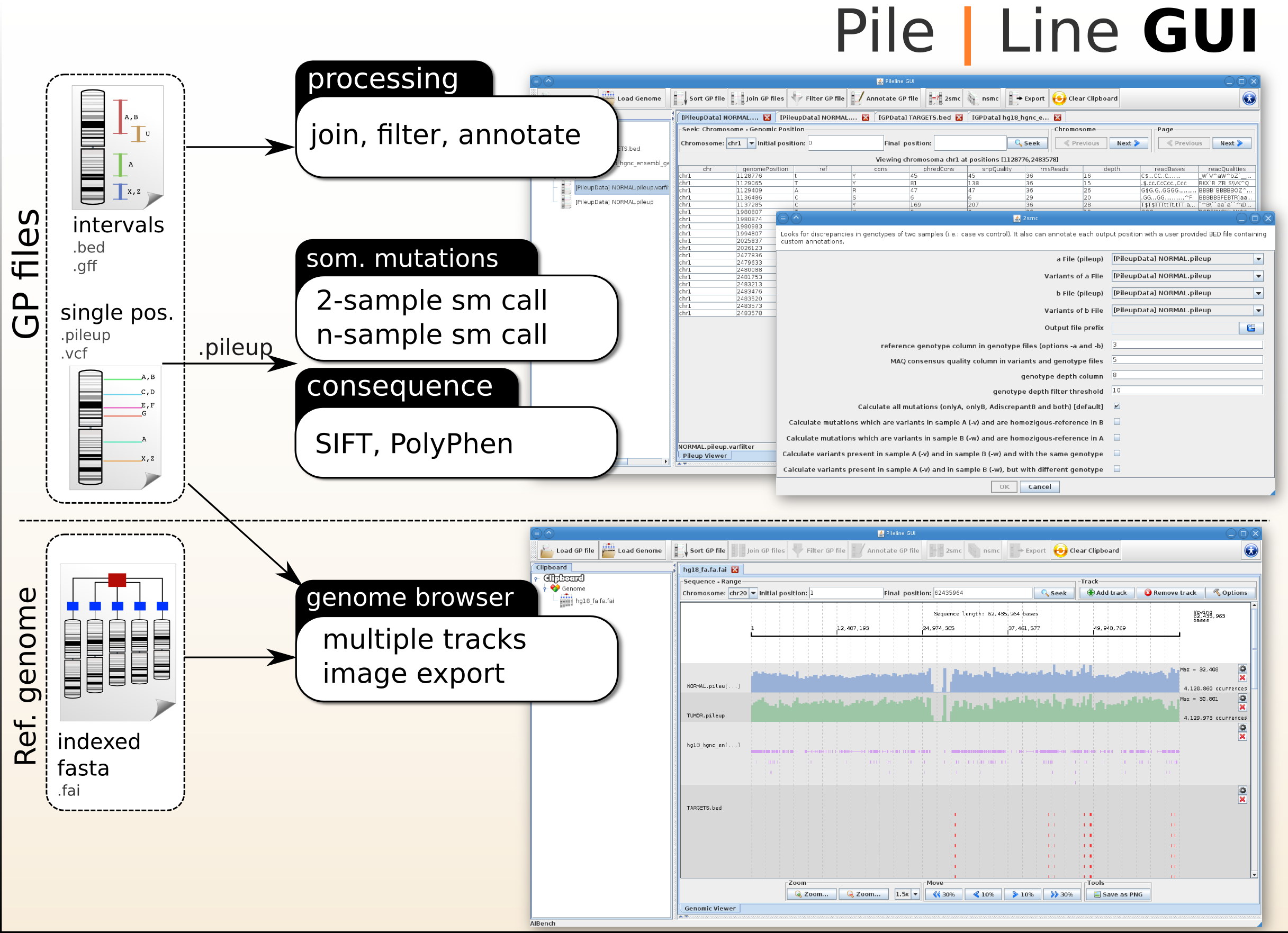
PileLine GUI includes a front-end of the PileLine toolkit, plus a genome browser.
Main Features
- Quick filtering and search within GP files without indexing steps.
- GP files comparisons.
- Full annotation of GP files with human dbSNP, HGNC Gene Symbol and Ensembl IDs. Custom annotations are also allowed and may be supplied through standard .BED or .GFF files.
- SIFT, PolyPhen-2 and Firestar compatible inputs to facilitate the biological interpretation of huge lists of variants.
- Genotyping quality control functionality to estimate performance metrics (Harismendi et al. 2009) on detecting homo/heterozigote variants against a given gold standard genotype.
- Modular design to facilitate the inclusion of new functionalities.
Getting started
New to PileLine? Please, follow our Quick Start.
PileLine Commands
Processing and Annotation Commands
- pileline-fastseek
Prints a given range of a GP file.
- pileline-sort
Sorts GP files by coordinate.
- pileline-fastjoin
Joins two SORTED GP files.
- pileline-rfilter
Filters (or annotates) a positional file with range-based annotations (in bed format). Each position that is inside of a specific range is annotated.
- pileline-pileup2sift
Generates SIFT compatible infiles from pileup files.
- pileline-pileup2polyphen
Generates PolyPhen-2 compatible infiles from pileup files.
- pileline-pileup2firestar
Generates Firestar compatible infiles from GP files.
Analysis Commands
- pileline-2smc
Looks for discrepancies in genotypes of two samples (i.e.: case vs control). It also can annotate each output position with a user provided BED file containing custom annotations.
- pileline-nsmc
Compares n samples reporting consistent variants.
- pileline-genotest
Calculates the NGS performance on genotyping, surveying a set of genomic positions whose genotype is known in the sample. This functionality requires a previous step using *pileline-genindex command for genome indexing.
Use Cases

- Perform 2 samples comparison
pileline-2smc.sh –a <file_A.pileup> –b <file_B.pileup> –v <variants_file_A.pileup> –w <variants_file_B.pileup> –o <out.txt> -d <min_depth>
- Perform n samples comparison
pileline-nsmc.sh --a-samples<GPfile_a1>,<GPfile_a2>,<GPfile_a3> --b-samples <GPfile_b1>,<GPfile_b2>,<GPfile_b3>
- Sort GP files
pileline-sort.sh -i <input_GP_file.txt> -o <outfile.sorted.txt>
- Annotate a GP file with dbSNP
pileline-fastjoin.sh -a <GP_file.txt> -b dbSNP130.txt --left-outer-join
- Annotate a GP file with genes
pileline-rfilter.sh --annotate -A <GP_file.txt> -b <genes.bed>
- Filter pileup to exon loci
pileline-rfilter.sh -A <GP_file.txt> -b <exons.bed>
- Generate column compatible to SIFT intput
pileline-pileup2sift.sh -i <file.pileup>
- Perform a genotyping test for quality control
# Warning: Check that your alleles in the <gold_genotype.sorted> file are expressed in the same strand as the # reference genome sequence used in your NGS experiment. Typically forward (+) strand. ## Step1. #Create reference index <ref_genome.pileline> using pileline-genindex command. pileline-genindex --index -i <ref_genome.pileline> -g <ref_genome.fa> ## Step2. #Create genotest file (required). pileline-genotest --create-genotest-file <experiment.genotest> -p <GP_file.txt> -g <gold_genotype.sorted> -r <ref_genome.pileline> ## Step3. QC analysis. #Generate all performance metrics for several thresholds pileline-genotest -a <experiment.genotest> --batch-t 0,255,1 #Generate values for ROC curve plot (outfile compatible to ROCR R package) pileline-genotest -a <experiment.genotest> --roc #Generate a metrics table of performance at a given threshold. pileline-genotest -a <experiment.genotest> -t <snpq_treshold>
PileLine GUI
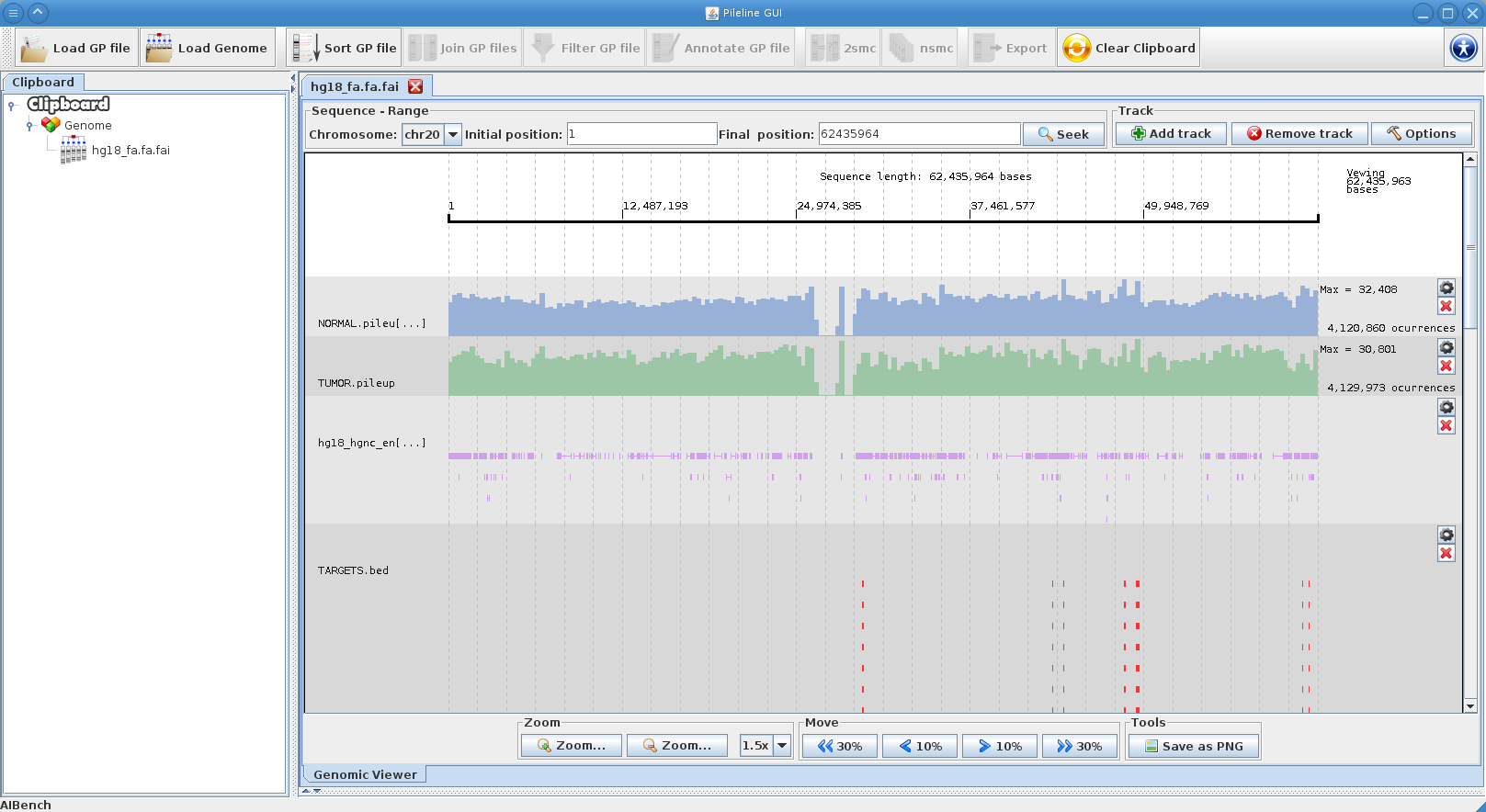
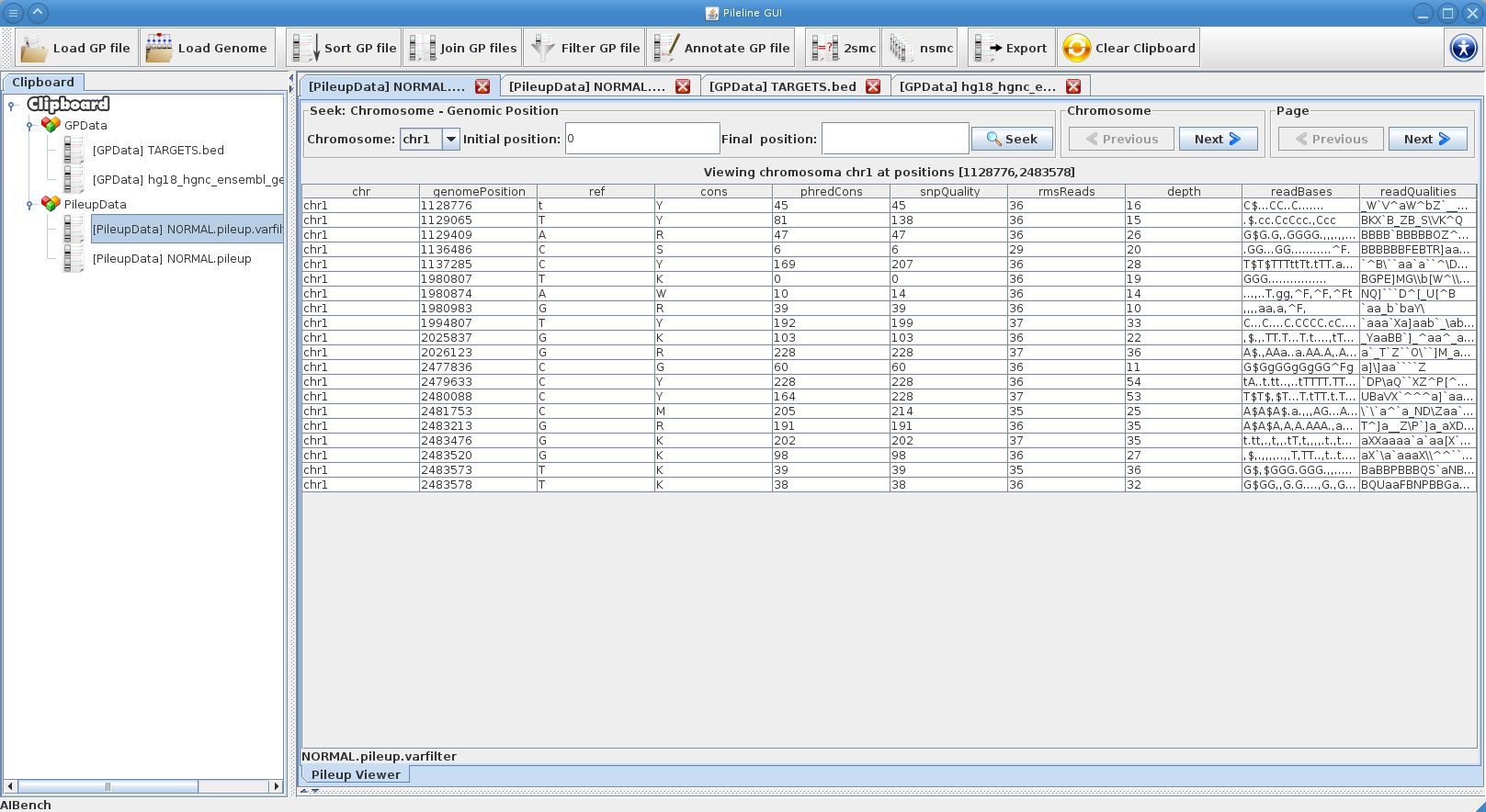
PileLine GUI is a front-end of the PileLine toolkit, plus a genome browser. With this intuitive graphical desktop application you can run the following tasks:
- Processing commands of GP files, like seek, join, annotate and filtering.
- Perform 2-samples and n-samples point somatic mutation calling (via the PileLine 2smc and nsmc commands).
- Browse GP files in a interactive local genome browser.
You can download PileLine GUI from Downloads.